TUTORIAL
The main menu provides a quick selection of the types of data to mine. Each data type is linked to several available studies ready to be explored. A table showing list of studies available will also have extensive information about the data represented in each study. (See figure 1)
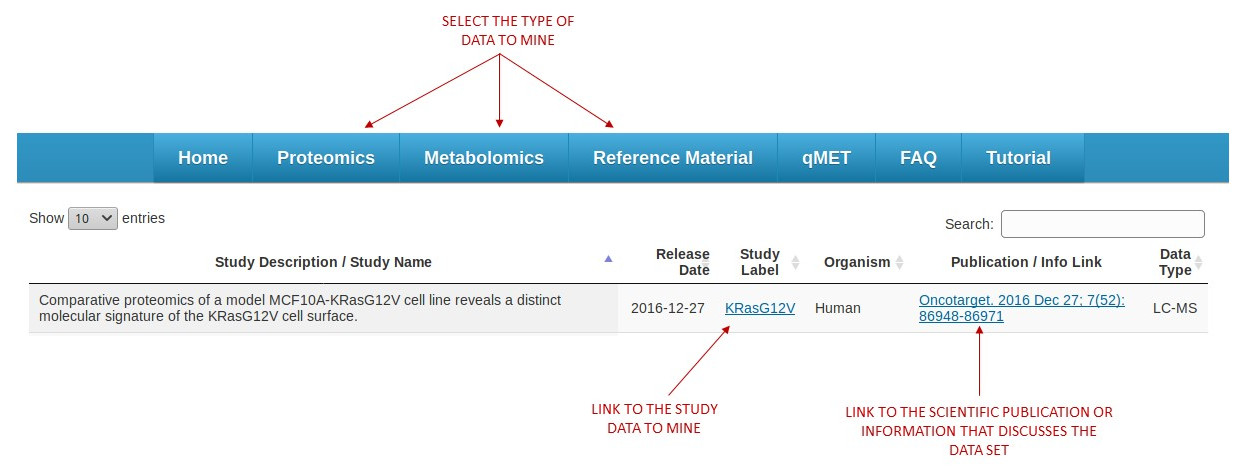 [figure 1]
[figure 1]Clicking on study label will bring an option to view experiments (runs) or identifications from the selected study. (See figures 2 and 3)
For an experiment-based search click on a file name to see the graphical representation of the experiment. (See figure 2)
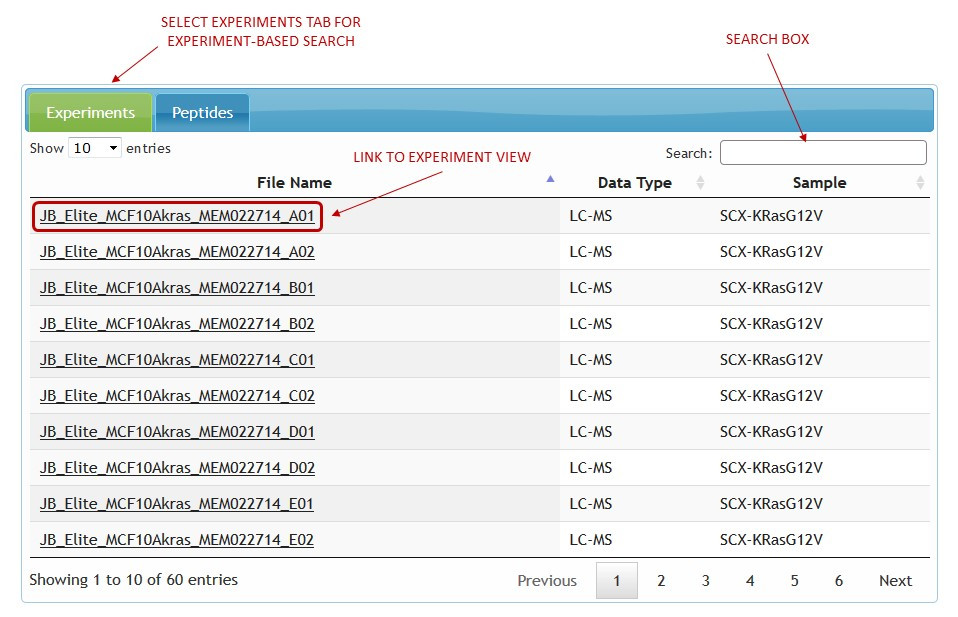 [figure 2]
[figure 2]For a study-based search select the identifications tab (Peptides, Metabolites etc.) and click on identification link to view spectrum representation and metadata. (See figure 3). Identifications can be filtered by searching the entire table content using the search box in the top right corner. Using search boxes at the footer of the table, filtering can be done by searching on any single table column as well as a combination of columns.
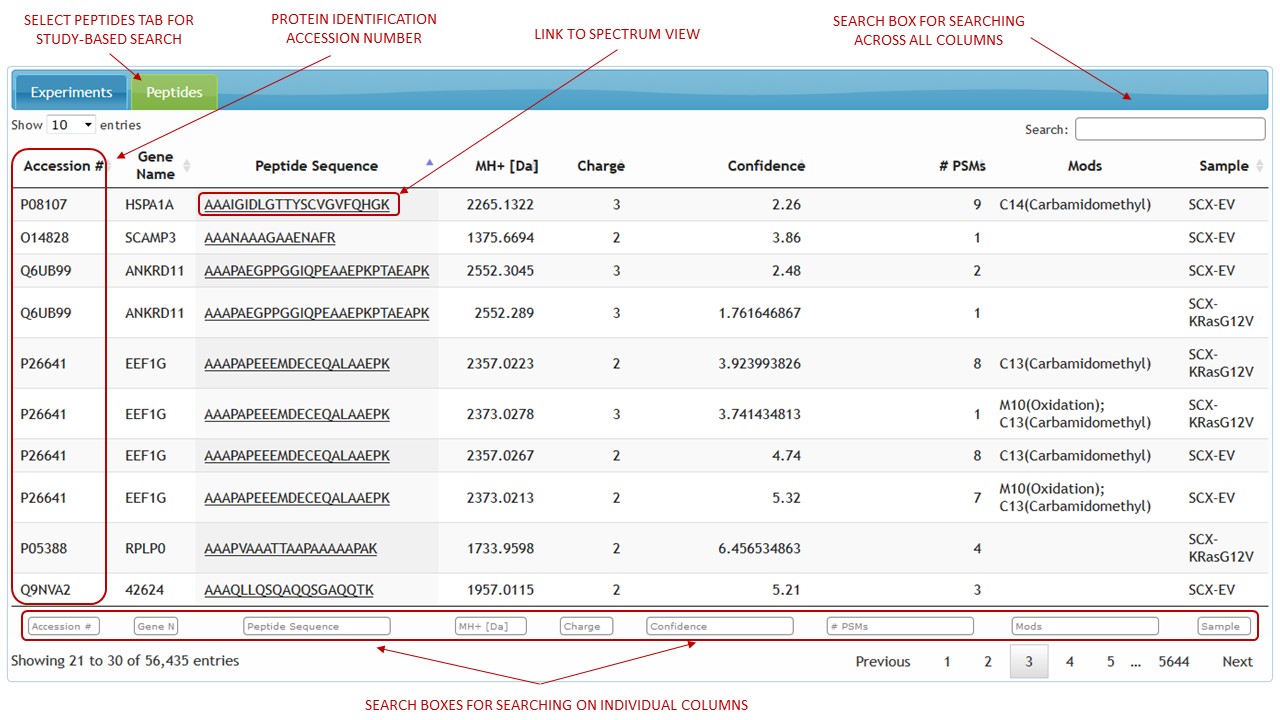 [figure 3]
[figure 3]Experiment-based search:
Experiment view:
The experiment view shows chromatogram, with identification points. On the right side of the chromatogram is an identification table with two columns displaying the identification title and the retention time. The table can be searched for both the identification title and the retention time as well as sorted on any of the two columns. To sort the table on any given column, click on the column header once for ascending order and twice for descending order. Purple arrow next to column header indicates the sorting order. (See figure 4)
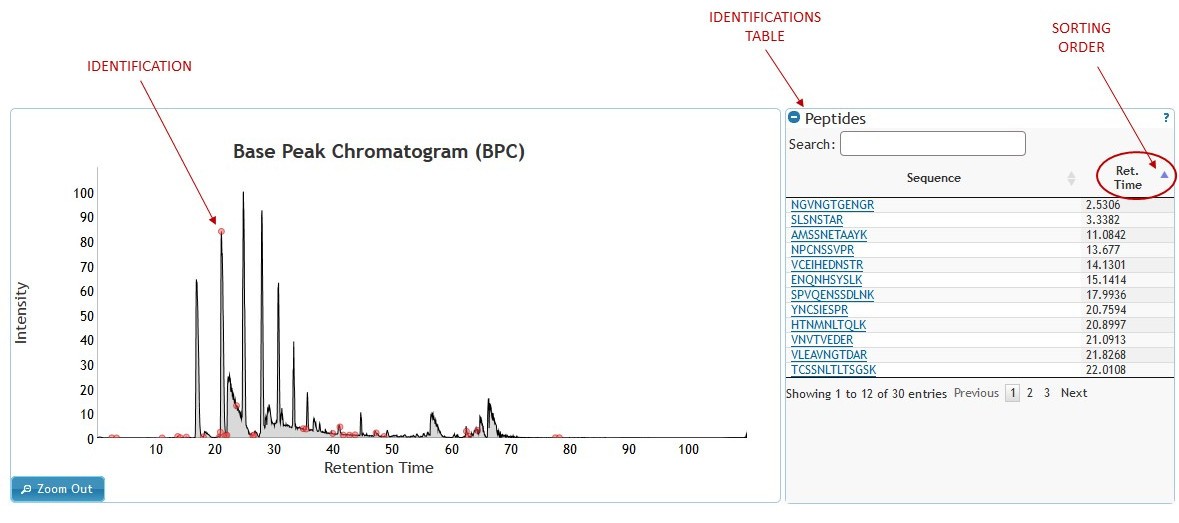 [figure 4]
[figure 4]Chromatogram Zooming:
Zooming in on an area of a given chromatogram can be done by clicking on any point on the chromatogram and dragging the mouse across the graph while holding down the mouse button. The selected area will be highlighted in gray. After releasing the mouse button the graph will zoom in to the selected area. To zoom out click on the "Zoom Out" button. (See figure 5)
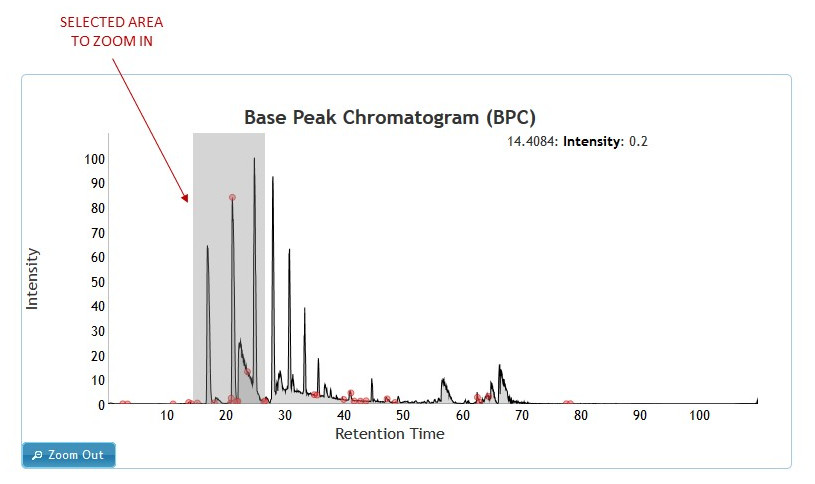 [figure 5]
[figure 5]Viewing Spectra:
Hovering with the mouse over the chromatogram will show a black dot on the chromatogram. The dot will trace the mouse pointer as it moves across the graph. (See figure 6)
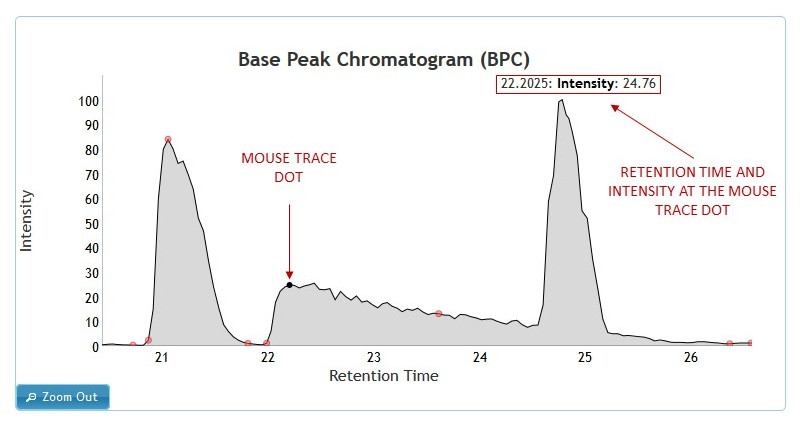 [figure 6]
[figure 6]Clicking on any point on the chromatogram will bring up Ms1 spectrum associated with the retention time. In case of LC/MS data, MS2 Spectra tab will be available to access and view the MS2 level spectra.
Just like the chromatogram, the spectrum can be zoomed in by dragging the mouse across the graph while holding down the mouse button and releasing it once the desired area has been highlighted. Click once on the spectrum viewer to zoom out to full spectrum view. (See figure 7)
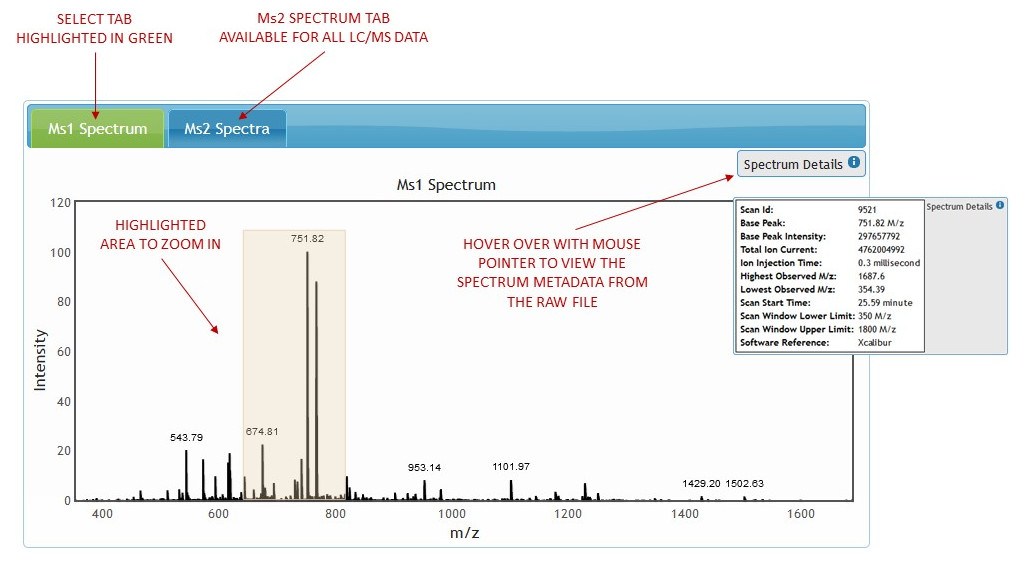 [figure 7]
[figure 7]Hover the mouse pointer over "Spectrum Details" box to expand it and view Ms1 Spectrum metadata from the raw file. (See figure 8)
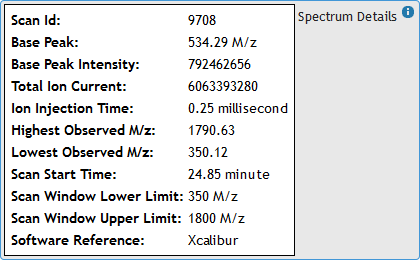 [figure 8]
[figure 8]Moving the mouse away from the box will automatically hide the metadata.
Clicking on the MS2 Spectra tab will load the first fragment ion spectrum. (See figure 9)
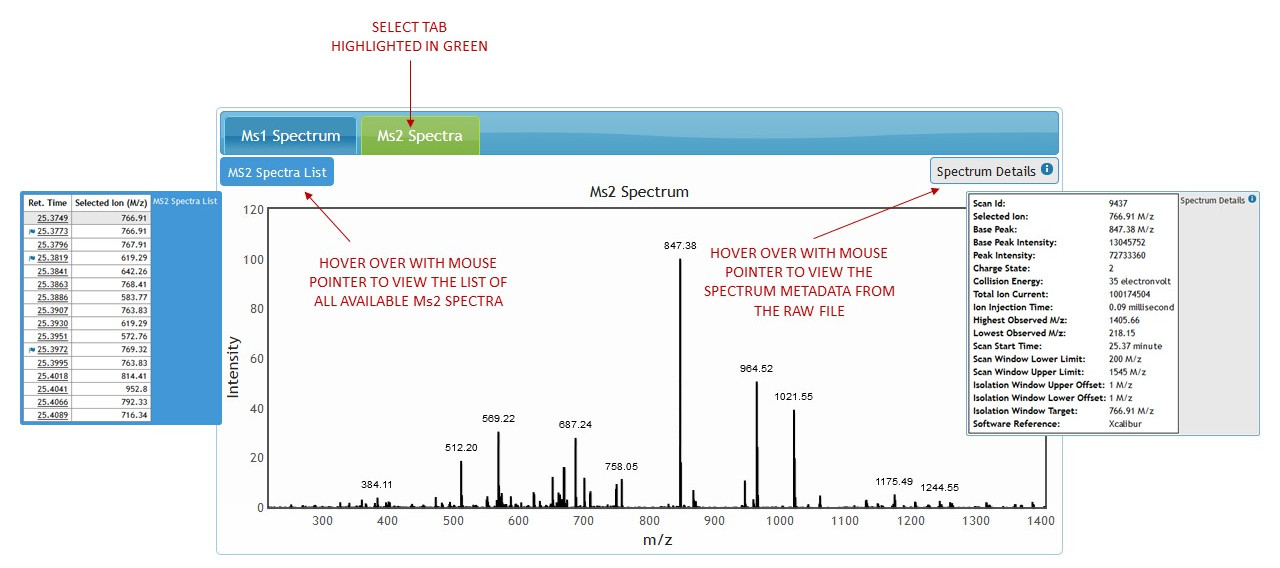 [figure 9]
[figure 9]To view links for other available fragment ion spectra hover mouse over "MS2 Spectra List" box to expand it. (See figures 9 and 10)
Gray row indicates currently selected spectrum. Clicking on retention time will load a new MS2 Spectrum. Blue flags next to retention time indicate that the fragmentation ion has been identified. These identifications are also indicated by red marks on the chromatogram graph and can also be found in the identifications table next to the chromatogram graph. Moving the mouse away from the box will automatically hide the spectra links. (See figure 10)
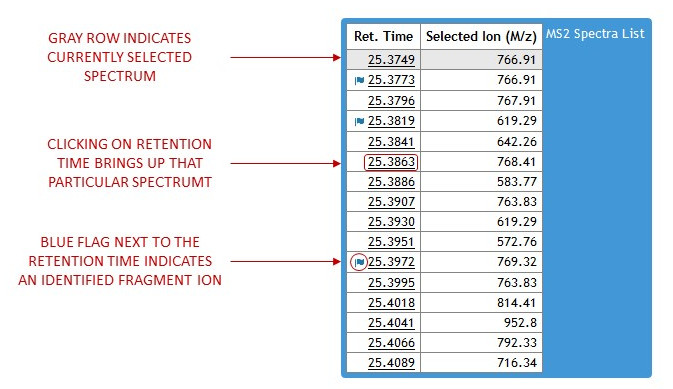 [figure 10]
[figure 10]Hover the mouse pointer over "Spectrum Details" box to expand it and view MS2 Spectrum metadata from the raw file. (See figures 9 and 11)
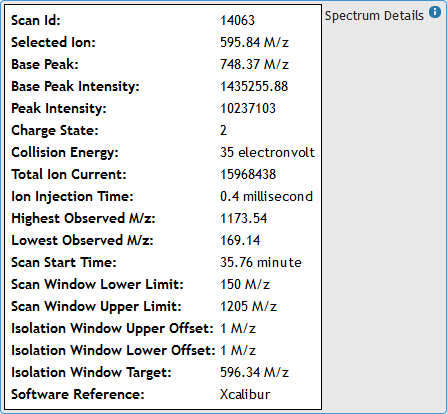 [figure 11]
[figure 11]Moving the mouse away from the box will automatically hide the metadata.
Viewing identifications:
There are two ways to quickly view only identifications in the experiment view. (See figure 12)
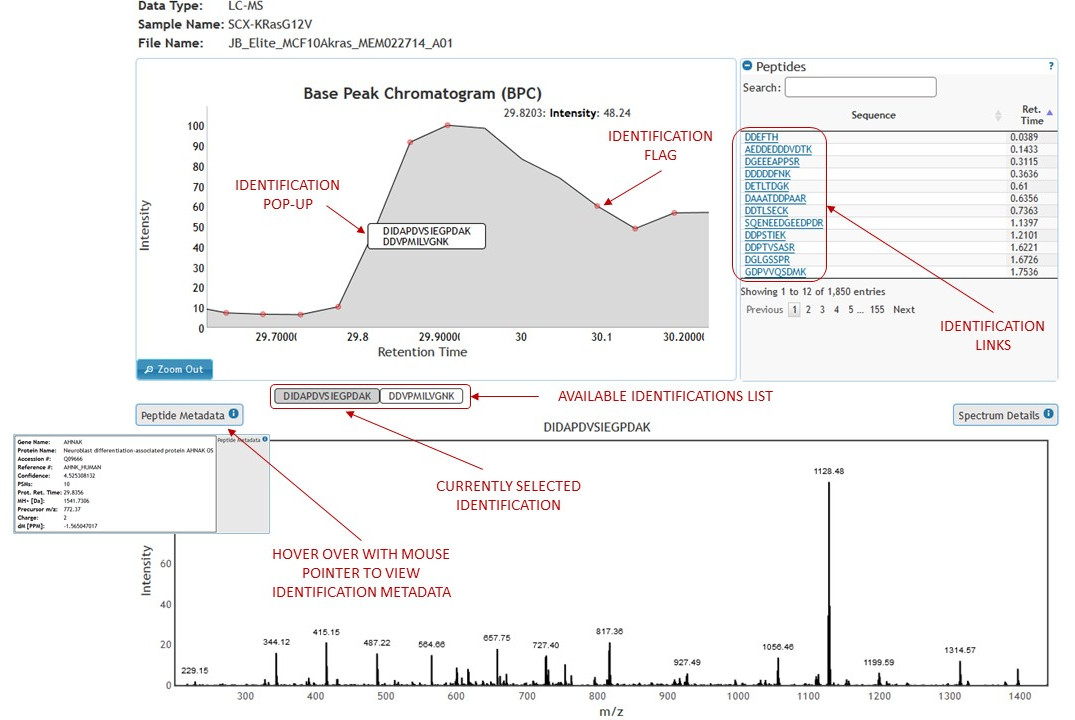 [figure 12]
[figure 12]Identification metadata can be viewed by hovering mouse over the metadata box. (See figures 12 and 13)
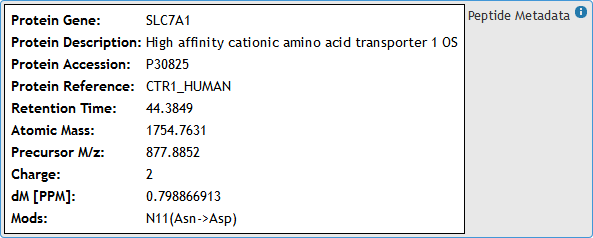 [figure 13]
[figure 13]Moving the mouse away from the box will automatically hide the metadata.
Study-based search:
Identifications can be searched across the database through the identifications table. (See figure 14)
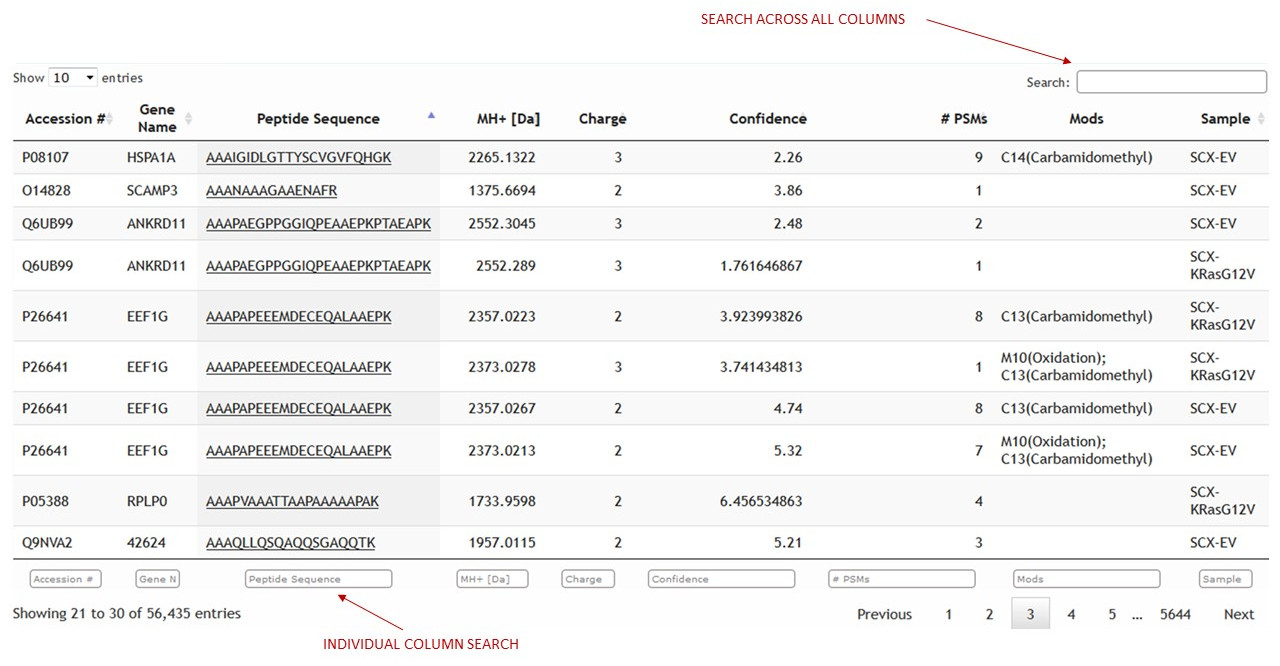 [figure 14]
[figure 14]The search can be across all columns or across individual columns at the bottom of the identifications table.
Clicking on an identification will bring up the spectrum view. (See figure 15)
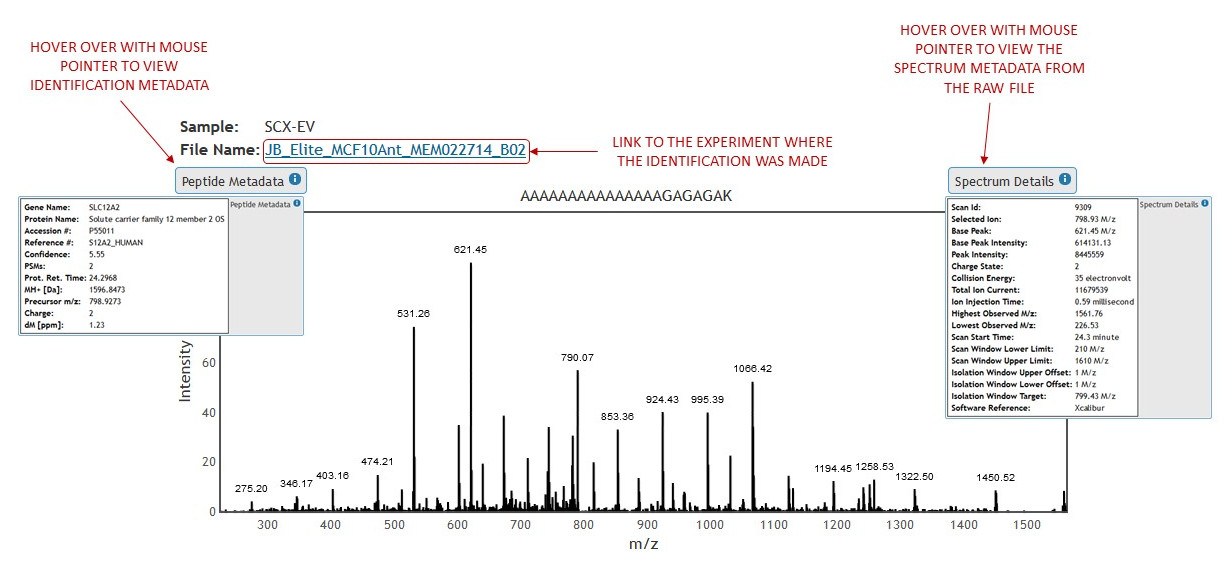 [figure 15]
[figure 15]In the top left corner of the spectrum view are the details showing the data set name from which the spectrum was taken and the link to the experiment view of the data set that the identification belongs to.
Hovering over identification and spectrum detail boxes will expand identification meta data and raw spectrum data respectively (See figures 13 and 11)